Analysis of Lignins Using 31P Benchtop NMR Spectroscopy
As a class of complex cross-linked phenolic macromolecules, lignin is an important component of plant-based biomass.1 Biologically, lignin is a major component of lignocellulose, helping to strengthen cell walls in wood. A by-product of paper production, most of the lignin continues to be burned for energy production, which is a shame considering its monomeric sub-units are important building blocks commonly obtained from petroleum feedstocks.2
In Canada, researchers have been trying to find value-added uses for lignin for years, with several success stories emerging more recently. Of note, the West Fraser plant in Hinton, Alberta, can currently produce 10,500 tones of lignin per year. This plant uses the LignoForce System™ jointly developed by FPInnovations and NORAM Engineering and has allowed for the production of lignin for specific use as a plywood adhesive.
More recently still, lignin is being explored as a possible replacement for bitumen in asphalt. In Europe, tests have confirmed that up to 50% of the bitumen can be replaced by lignin. In Canada, however, testing is currently underway to confirm whether the harsh Canadian Winters can have an impact on the durability of these new paved roads. As of late 2021, 3 road segments using lignin-containing asphalt have been paved in Canada and are actively being studied for their overall performance: Sturgeon County, Alberta (August 2021); Thunder Bay, Ontario (September 2021); Québec City, Québec (October 2021).
The bitumen market for asphalt in Canada is currently estimated at ~4M tonnes per year. While our road network adds up to hundreds of thousands of kilometers, it is estimated that only ~40% of the total network is paved. As this network continues to grow and as these roads require maintenance, increases in paving requirements are expected. To help with this, a sustainable and renewable resource such as lignin will be crucial as we collectively aim to rely less on petroleum feedstocks.
Because lignin is such a complex polymer (Figure 1), its characterization is not straightforward. Each lignin sample is unique and contains different amounts of various sub-units. These amounts are important to fully understand and predict how a lignin sample will behave chemically and physically.
Figure 1. Representation of various repeating substructures of lignin, a complex cross-linked phenolic polymer. Adapted from Argyropoulos et al.3
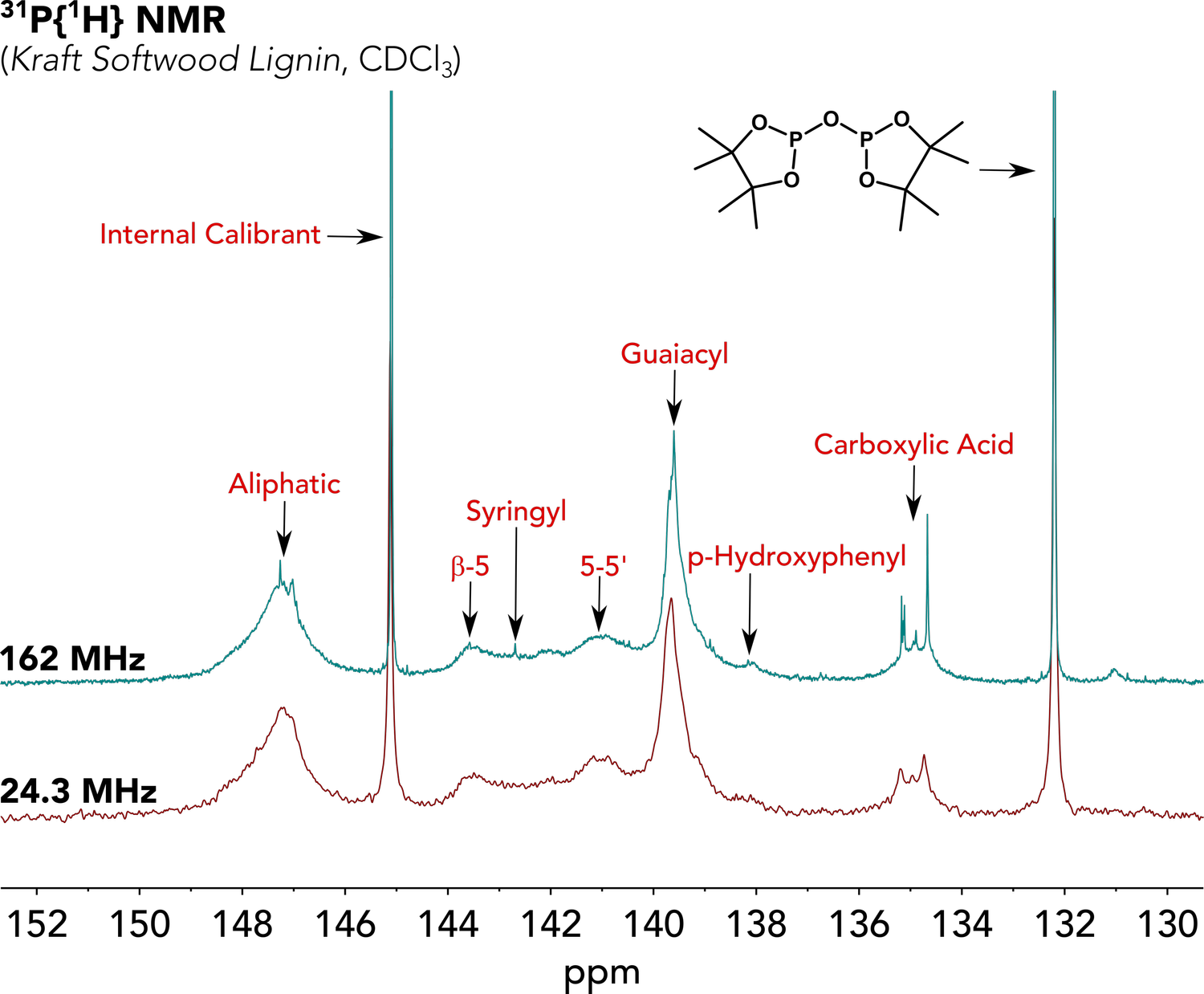
While 1H/13C NMR analyses tend to suffer from too much overlap when working with lignin, the use of a phosphitylating agent (e.g., TMDP) allows for the derivatization of every -OH group in the polymer with a phosphorus-containing group.3 Importantly, this allows for the 31P analysis of lignin samples (Figure 2). As an NMR nucleus, 31P is 100% naturally abundant, has a large chemical shift range (which is sensitive to the chemical environment), and gives rise to sharp lines. We have recently published a manuscript detailing our work on the quantitative assessment of lignin substructures using this phosphitylation approach, wherein we compare the data obtained on a 60 MHz benchtop NMR instrument to a traditional 400 MHz high-field instrument. Our results match very closely, indicating that benchtop NMR technology is a powerful tool for the analysis of lignins!
Figure 2. Stacked 31P (24.3 MHz, bottom; 162 MHz, top) NMR spectra of a phosphitylated kraft softwood lignin, with some key chemical shift regions identified.
If this topic interests you, please read our manuscript for the full details of this study. If you have any questions about the incorporation of benchtop NMR into your lignin workflows, please don’t hesitate to contact us!
References
(1) Ragauskas, A. J.; Williams, C. K.; Davison, B. H.; Britovsek, G.; Cairney, J.; Eckert, C. A.; Frederick, W. J.; Hallett, J. P.; Leak, D. J.; Liotta, C. L.; Mielenz, J. R.; Murphy, R.; Templer, R.; Tschaplinski, T. Science 2006 , 311, 484–489.
(2) Li, Y.; Shuai, L.; Kim, H.; Motagamwala, A. H.; Mobley, J. K.; Yue, F.; Tobimatsu, Y.; Havkin-Frenkel, D.; Chen, F.; Dixon, R. A.; Luterbacher, J. S.; Dumesic, J. A.; Ralph, J. Sci. Adv. 2018 , 4, 1–10.
(3) Meng, X.; Crestini, C.; Ben, H.; Hao, N.; Pu, Y.; Ragauskas, A. J.; Argyropoulos, D. S. Nat. Protoc. 2019, 14, 2627–2647.